Concentrated solutions of single chain nanoparticles: a simple model for intrinsically disordered proteins under crowding conditions
Single-chain nanoparticles (SCNPs) are an emergent class of soft nano-objects with promising applications in e.g., nanomedicine, biosensing, bioimaging, or catalysis. They are synthesized through intramolecular cross-linking of polymer precursors. In this work, the research team performed a detailed simulation analysis of the conformations of SCNPs, revealing that they share basic ingredients with intrinsically disordered proteins (IDPs), as topological polydispersity, compact domains and sparse regions.
IDPs are highly abundant in eukaryotes. Their biological function is founded on their internal dynamics and flexibility, enabling them to respond quickly to environmental changes and to bind with different cellular targets. As a direct consequence of their malleability, the structural, dynamic and associative properties of IDPs can be affected by macromolecular crowding in vivo, substantially differing from the observations in vitro at highly dilute conditions.
The research team exploited the structural analogies between SCNPs and IDPs and investigated the conformational properties of SCNPs in concentrated solutions. SCNPs provide a model system that shares universal structural features with IDPs and is free of specific interactions, allowing to investigate separately the purely steric excluded-volume contributions to crowding. For this purpose, large-scale simulations of a generic bead-spring model for solutions of SCNPs and small-angle neutron scattering (SANS) experiments on real systems were combined, covering the whole concentration range from infinite dilution to melt density. The role of the internal degree of disorder of the SCNP on its collapse behavior under macromolecular crowding was analyzed.
It was found that crowding leads to collapsed conformations of SCNPs resembling those of the crumpled globular class. This behavior was already found at volume fractions (about 30 %) that are characteristic of crowding in living cells.
The results in SCNPs propose a universal scenario for IDPs: steric crowding in cell environments lead IDPs to adopt crumpled globular conformations. The usual transition from self-avoiding to random coil (Gaussian) conformations found in linear macromolecules is only a particular case, taking place in the limit of fully disordered IDPs.
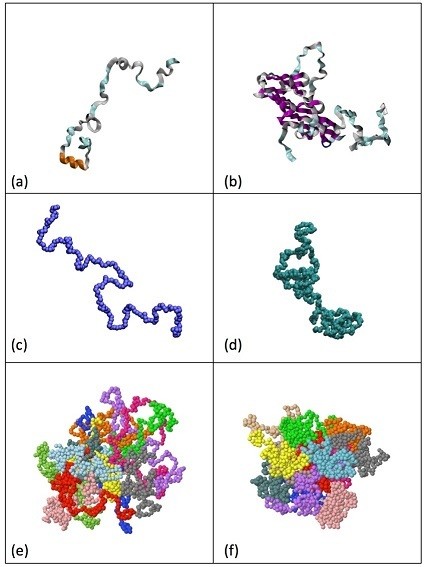
Figure: (a): IDP in the limit of full disorder. (b): IDP with a compact domain. (c): Polymer chain. (d): SCNP. (e): Concentrated solution of polymer chains, showing Gaussian conformations. (f): Concentrated solution of SCNPs, showing crumpled globular conformations.