MOLECULAR ARCHITECTURE OF SINGLE-CHAIN NANOPARTICLES
Single-chain nanoparticles (SCNPs) are unimolecular soft nano-objects, consisting of individual polymer chains collapsed to a certain degree by means of intramolecular bonding.
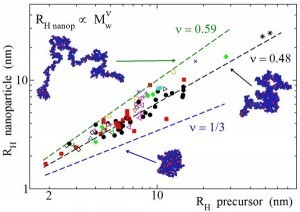
Many of the potential applications of SCNPs rely on their particular molecular architecture. Even if the ultimate goal is to produce globular protein-like soft nanoparticles, recent small-angle neutron scattering (SANS) and small-angle X-ray scattering (SAXS) results—supported by computer simulations—indicate that SCNPs in solution actually adopt sparse configurations. Herein we compile size data from the literature for a large number of SCNPs in solution, covering from covalent to noncovalent bonded SCNPs, and provide a comparison with the corresponding data for compact or partially swollen globules of the same nature and molar mass. This comparison gives a clear idea of how far from the compact globule limit are current SCNPs. A quantification of the departure from the globular state is provided in terms of size scaling laws. This procedure facilitates a comparison with the size scaling laws observed for folded proteins with globular conformation as well as intrinsically disordered proteins which, on average, exhibit a certain local compaction when compared to chemically denatured proteins. Lastly, the underlying physical mechanism for the noncompact morphology of SCNPs in solution is put forward, and guidelines for the potential synthesis of true SCNP globules in solution are suggested.
You will find more information in: ACS Polymer Science Audio/Podcast (Interview with Managing Editor, Paulomi Majumder, from min. 8:05 to 15:32)